Management of Patients With a Genetic Variant of Unknown Significance
Accurate risk assessment of developing cancer often includes genetic testing for germline mutations, which has clinical and treatment implications for the patient and his or her family members. When a mutation is detected, aggressive measures for cancer prevention and detection are often implemented. Depending on the gene or genes tested, a variable percentage of patients will receive a test report stating that a variant of unknown significance (VUS) has been detected. This means that a change in the genetic sequence has occurred, but whether the change is associated with an increased risk of cancer or another disease is unclear or unknown. The results are confusing and noninformative. Oncology nurses will undoubtedly encounter patients with a VUS; consequently, they need to understand the controversies and ambiguities that surround these test results.
Jump to a section
Accurate risk assessment of developing cancer often includes genetic testing for germline mutations, which has clinical and treatment implications for the patient and his or her family members. When a mutation is detected, aggressive measures for cancer prevention and detection are often implemented. Depending on the gene or genes tested, a variable percentage of patients will receive a test report stating that a variant of unknown significance (VUS) has been detected. This means that a change in the genetic sequence has occurred, but whether the change is associated with an increased risk of cancer or another disease is unclear or unknown. The results are confusing and noninformative. Oncology nurses will undoubtedly encounter patients with a VUS; consequently, they need to understand the controversies and ambiguities that surround these test results. Resources that may be offered by healthcare providers and aid in patient understanding of VUS results are available (see Figure 1).
Scope of the Problem
Several possibilities follow the reporting of genetic testing results, including a known pathogenic or deleterious mutation, a VUS, or a negative report (see Table 1). A negative test result does not necessarily indicate that the patient has no increased risk for developing cancer. A true negative occurs when a known pathogenic mutation is already present in the family. If the patient tests negative, he or she has not inherited the elevated risk but still has the population risk for developing cancer. When a negative test result occurs in the first person tested in the family (no prior known mutation exists), no pathogenic mutation has been detected in that person, and the result is noninformative. The individual may have benign polymorphisms that most likely will not be reported. Alternatively, he or she may not have a germline mutation or may have a germline mutation for which testing is not available. In addition, the cause of cancer could be attributed to another gene or hereditary syndrome. 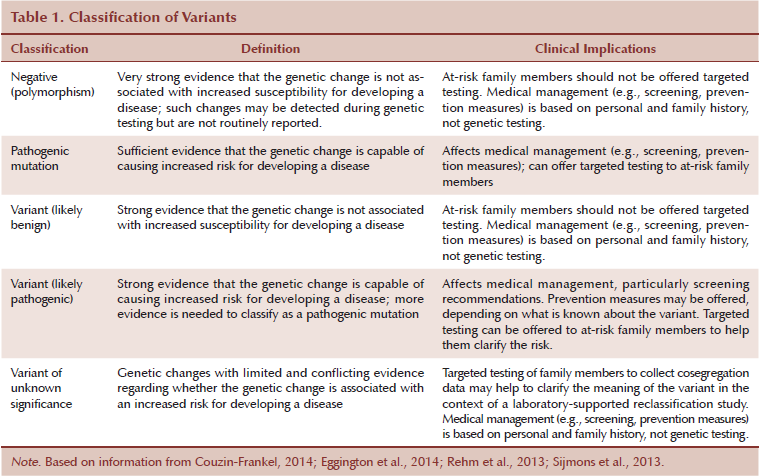
Next-generation sequencing has resulted in the detection of pathogenic mutations in less common genes and has increased the number of VUSs found because more genes are included in the analyses. Less is known about some of the newer genes (e.g., their clinical implications, whether genetic changes within newer genes are pathogenic). For example, Myriad Genetic Laboratories reported that the VUS rate decreased from 12.8% of all BRCA1 and BRCA2 test results in 2002 to 2.1% of all test results in 2013, and credited the decline to more being learned about the genes and the existence of a larger database (Eggington et al., 2014). In a study of 175 patients who underwent next-generation panel testing, 428 VUSs were identified in 39 different genes, resulting in an average of 2.1 VUSs per patient (Kurian et al., 2014). Rates of VUSs in next-generation panels may approach 20% (Selkirk et al., 2014). Variant rates may be higher in minority populations because they are not represented as well in databases (Murray, Cerrato, Bennett, & Jarvik, 2011). Many variants are reclassified as more information becomes available, but, in many cases, this can take years (Cheon, Mozersky, & Cook-Deegan, 2014).
Reclassification Strategies
Multiple approaches to VUS classification and reclassification exist, but no universally accepted standard or approach for determining if a variant is pathogenic or how a VUS should be reported or reclassified is available (Cheon et al., 2014). Some databases share data about variants, but a great deal of data about variants are proprietary. For example, Myriad Genetic Laboratories had a patent on BRCA1 and BRCA2 testing until 2013; consequently, the company has a large proprietary database of BRCA1 and BRCA2 data, which has enabled its reclassification of many VUSs in BRCA1 and BRCA2 (Cheon et al., 2014). The International Society for Gastrointestinal Hereditary Tumours (InSiGHT) has demonstrated how a collaborative effort to have a central repository for variant classification can be effective. InSiGHT examined variants in Lynch syndrome for the MLH1, MSH2, MSH6, and PMS2 genes, resulting in the evaluation of 2,360 variants and the reclassification of 605 variants (Thompson et al., 2014). The best solution would be to have a central repository for all data, but that is not yet a reality.
Multiple strategies are often combined and required to determine if a VUS is pathogenic or benign (Eggington et al., 2014; Millot et al., 2012; Moghadasi et al., 2013; Radice, De Summa, Caleca, & Tommasi, 2011; Rehm et al., 2013; Sijmons, Greenblatt, & Genuardi, 2013). Most laboratories use a variety of methods to reclassify a variant (see Figure 2). Reclassification is not universally carried out the same way across laboratories and agencies.
Implications for Patient Care
The noninformative nature of a VUS leads to confusion and challenges for patients, families, and healthcare providers. When a VUS is reported, no clear information regarding whether the patient is at higher risk for developing cancer is available. A very real concern is that the detection of a variant will increase anxiety and possibly lead to overtreatment (Kurian et al., 2014). Unfortunately, many variants will not be reclassified quickly enough to help with clinical decision making (Murray et al., 2011). Patients’ making irreversible treatment decisions (e.g., prophylactic surgery) based on a report with a VUS that is later reclassified as benign is of particular concern (Murray et al., 2011). This can be devastating in women who undergo prophylactic mastectomies or premenopausal women who undergo oophorectomy. Healthcare providers may also incorrectly interpret the meaning of a VUS. In one small study of 24 healthcare providers who were queried on the management of a patient who had received a VUS result, all 24 reported incorrectly that they would refer a sibling of a patient with a VUS for predictive genetic testing (Richter et al., 2013).
The psychosocial impact of a VUS may be substantial (Richter et al., 2013). Patients with lower education levels are at increased risk for misunderstanding the meaning of a VUS. In a study of 36 women with a BRCA VUS who had received genetic counseling, 30% mistakenly believed that the VUS increased their cancer risk, and worry about their cancer risk increased after testing.
Pretest counseling, particularly when testing involves the ordering of next-generation panel testing, should include a clear discussion regarding the very real possibility of detecting a VUS and the noninformative nature of such a result. The possibility of a VUS is included in the consent forms that patients often sign prior to testing, which offer an opportunity to reiterate this information.
When a VUS is reported, the first step a genetics professional typically takes is to contact the laboratory to gather as much information as possible about the variant and determine if the family is eligible for family studies. Many times, if another family member is diagnosed with cancer, the laboratory will offer testing to that individual to help clarify if the variant tracks with cancer. In some cases, families also qualify for enrollment in research studies, such as the Prospective Registry of Multiplex Testing, which is a joint effort of academic medical centers and commercial laboratories to learn more about variants (Couzin-Frankel, 2014).
During disclosure of a VUS, the patient and family must understand that the result is noninformative; this can be stressful and confusing. Recommendations for cancer prevention and surveillance must then be made based on personal risk factors and the family history for the proband and family. Testing of at-risk family members is not recommended when a VUS is identified. These points should be communicated to the patient verbally and in a recommendations letter summarizing what was discussed during the disclosure visit.
When variants are reclassified, the responsibility to share the information with the patient belongs to either the laboratory or genetics professional, but it typically falls to the latter. Recontacting patients is often more difficult than anticipated and can be time consuming (Cheon et al., 2014). Patients may move or die; in those cases, determining how to reach the patient or with whom to share results, respectively, is unclear. Many genetics professionals recommend that patients recontact them every 12 months to see if changes have occurred in what is known about a mutation or variant. Some patients will do this, but many will not. Patients should also be instructed to recontact the counselor if their personal or family history changes. Other family members may become eligible for family studies, and this new information about the family may aid in the reclassification of the variant.
Conclusion
Genetic testing that results in a VUS can be confusing and stressful for patients and healthcare providers. The very real possibility of a VUS should be discussed in pretest counseling, and patients should be informed that if a VUS is identified, recommendations will be based on personal and family history. Other at-risk family members will not be offered testing outside of a reclassification research effort. Reclassification can take years. Patients should be offered the opportunity to participate in studies or registries to facilitate reclassification. As VUSs are reclassified, patients should be informed, and recommendations for cancer prevention and surveillance should be modified based on the reclassification.
References
Cheon, J.Y., Mozersky, J., & Cook-Deegan, R. (2014). Variants of uncertain significance in BRCA: A harbinger of ethical and policy issues to come? Genome Medicine, 6, 121.
Couzin-Frankel, J. (2014). Unknown significance. Science, 346, 1167–1170.
Eggington, J.M., Bowles, K.R., Moyes, K., Manley, S., Esterling, L., Sizemore, S., . . . Wenstrup, R.J. (2014). A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clinical Genetics, 86, 229–237.
Kurian, A.W., Hare, E.E., Mills, M.A., Kingham, K.E., McPherson, L., Whittemore, A.S., . . . Ford, J.M. (2014). Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. Journal of Clinical Oncology, 32, 2001–2009.
Millot, G.A., Carvalho, M.A., Caputo, S.M., Vreeswijk, M.P., Brown, M.A., Webb, M., . . . Monteiro, A.N. (2012). A guide for functional analysis of BRCA1 variants of uncertain significance. Human Mutation, 33, 1526–1537.
Moghadasi, S., Hofland, N., Wouts, J.N., Hogervorst, F.B., Wijnen, J.T., Vreeswijk, M.P., & van Asperen, C.J. (2013). Variants of uncertain significance in BRCA1 and BRCA2 assessment of in silico analysis and a proposal for communication in genetic counselling. Journal of Medical Genetics, 50, 74–79.
Murray, M.L., Cerrato, F., Bennett, R.L., & Jarvik, G.P. (2011). Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: Variant reclassification and surgical decisions. Genetics in Medicine, 13, 998–1005. doi:10.1097/GIM.0b013e318226fc15
Radice, P., De Summa, S., Caleca, L., & Tommasi, S. (2011). Unclassified variants in BRCA genes: Guidelines for interpretation. Annals of Oncology, 22(Suppl. 1), i18–i23. doi:10.1093/annonc/mdq661
Rehm, H.L., Bale, S.J., Bayrak-Toydemir, P., Berg, J.S., Brown, K.K., Deignan, J.L., . . . Lyon, E. (2013). ACMG clinical laboratory standards for next-generation sequencing. Genetics in Medicine, 15, 733–747. doi:10.1038/gim.2013.92
Richter, S., Haroun, I., Graham, T.C., Eisen, A., Kiss, A., & Warner, E. (2013). Variants of unknown significance in BRCA testing: Impact on risk perception, worry, prevention and counseling. Annals of Oncology, 24(Suppl. 8), viii69–viii74. doi:10.1093/annonc/mdt312
Selkirk, C.G., Vogel, K.J., Newlin, A.C., Weissman, S.M., Weiss, S., Wang, C.H., & Hulick, P.J. (2014). Cancer genetic testing panels for inherited cancer susceptibility: The clinical experience of a large adult genetics practice. Familial Cancer, 13, 527–536. doi:10.1007/s10689-014-9741-4
Sijmons, R.H., Greenblatt, M.S., & Genuardi, M. (2013). Gene variants of unknown clinical significance in Lynch syndrome. An introduction for clinicians. Familial Cancer, 12, 181–187.
Thompson, B.A., Spurdle, A.B., Plazzer, J.P., Greenblatt, M.S., Akagi, K., Al-Mulla, F., . . . Genuardi, M. (2014). Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nature Genetics, 46, 107–115.
About the Author(s)
Suzanne M. Mahon, RN, DNSc, AOCN®, APNG, is a professor in the Department of Internal Medicine and in the School of Nursing at Saint Louis University in Missouri. No financial relationships to disclose. Mahon can be reached at mahonsm@slu.edu, with copy to editor at ONFEditor@ons.org.

